Notice: this Wiki will be going read only early in 2024 and edits will no longer be possible. Please see: https://gitlab.eclipse.org/eclipsefdn/helpdesk/-/wikis/Wiki-shutdown-plan for the plan.
Difference between revisions of "OHF STEM"
(reorg intro) |
(remove some out of date information) |
||
| Line 1: | Line 1: | ||
==STEM== | ==STEM== | ||
| − | ''' | + | What is '''Spatiotemporal Epidemiological Modeler''' ('''STEM''')? |
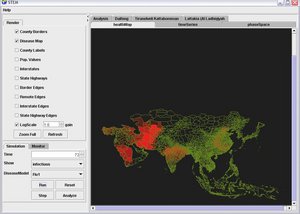
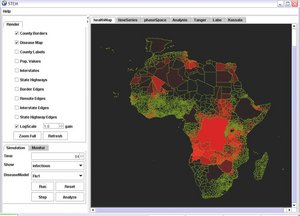
The Spatiotemporal Epidemiological Modeler (STEM) tool is designed to help scientists and public health officials create and use spatial and temporal models of emerging infectious diseases. These models could aid in understanding, and potentially preventing, the spread such diseases. | The Spatiotemporal Epidemiological Modeler (STEM) tool is designed to help scientists and public health officials create and use spatial and temporal models of emerging infectious diseases. These models could aid in understanding, and potentially preventing, the spread such diseases. | ||
| Line 8: | Line 8: | ||
'''How does it work?''' | '''How does it work?''' | ||
| − | The STEM application has built in Geographical Information System (GIS) data for every | + | The STEM application has built in Geographical Information System (GIS) data for almost every country in the world. It comes with data about country borders, populations, shared borders (neighbors), interstate highways, state highways, and airports. This data comes from various public sources. |
| − | STEM is designed to make it easy for developers and researchers to plug in their own models. It comes with spatiotemporal Susceptible/Infectious/Recovered (SIR) and Susceptible/Exposed/Infectious/Recovered (SEIR) models pre-coded with both deterministic and stochastic engines | + | STEM is designed to make it easy for developers and researchers to plug in their own models. It comes with spatiotemporal Susceptible/Infectious/Recovered (SIR) and Susceptible/Exposed/Infectious/Recovered (SEIR) models pre-coded with both deterministic and stochastic engines. |
| − | The parameters in any model are specified in XML configuration files. Users can easily change the weight or significance of various disease vectors (such as the weights of highways, shared borders, airports, etc). Users can also create their own unique vectors for disease. | + | The parameters in any model are specified in XML configuration files. Users can easily change the weight or significance of various disease vectors (such as the weights of highways, shared borders, airports, etc). Users can also create their own unique vectors for disease. Further details are available in the user manual and design documentation. |
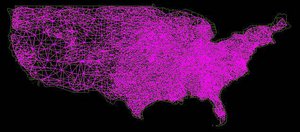
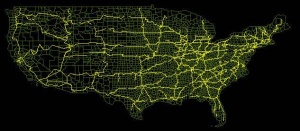
'''The U.S. Map in STEM with Admin 2 resolution showing county adjacency and interstate highways''' | '''The U.S. Map in STEM with Admin 2 resolution showing county adjacency and interstate highways''' | ||
| Line 21: | Line 21: | ||
[[Image:Stem4.jpg|300px]] | [[Image:Stem4.jpg|300px]] | ||
| − | The original version of [http://www.alphaworks.ibm.com/tech/stem STEM] was available for | + | The original version of [http://www.alphaworks.ibm.com/tech/stem STEM] was available for downloading on IBM's Alphaworks. It contained easy to follow instructions and many examples (various diseases and maps of the world). |
New developers who want to work on STEM II can find useful tools, conventions, and design information in the [[Welcome Developers]] article. | New developers who want to work on STEM II can find useful tools, conventions, and design information in the [[Welcome Developers]] article. | ||
| − | The STEM | + | The STEM code repository] will be hosted on the Eclipse OHF code repository. |
'''A recent publication on STEM:''' Ford, D.A., Kaufman, J.H., Eiron, I., "An extensible spatial and temporal epidemiological modeling system", | '''A recent publication on STEM:''' Ford, D.A., Kaufman, J.H., Eiron, I., "An extensible spatial and temporal epidemiological modeling system", | ||
| − | |||
International Journal of Health Geographics 2006, 5:4 [http://www.ij-healthgeographics.com/content/5/1/4 http://www.ij-healthgeographics.com/content/5/1/4] (17Jan2006) | International Journal of Health Geographics 2006, 5:4 [http://www.ij-healthgeographics.com/content/5/1/4 http://www.ij-healthgeographics.com/content/5/1/4] (17Jan2006) | ||
Revision as of 12:31, 29 January 2007
STEM
What is Spatiotemporal Epidemiological Modeler (STEM)?
The Spatiotemporal Epidemiological Modeler (STEM) tool is designed to help scientists and public health officials create and use spatial and temporal models of emerging infectious diseases. These models could aid in understanding, and potentially preventing, the spread such diseases.
Policymakers responsible for creating strategies to contain diseases and prevent epidemics need an accurate understanding of disease dynamics and the likely outcomes of preventive actions. In an increasingly connected world with extremely efficient global transportation links, the vectors of infection can be quite complex. STEM facilitates the development of advanced mathematical models, the creation of flexible models involving multiple populations (species) and interactions between diseases, and a better understanding of epidemiology.
How does it work? The STEM application has built in Geographical Information System (GIS) data for almost every country in the world. It comes with data about country borders, populations, shared borders (neighbors), interstate highways, state highways, and airports. This data comes from various public sources.
STEM is designed to make it easy for developers and researchers to plug in their own models. It comes with spatiotemporal Susceptible/Infectious/Recovered (SIR) and Susceptible/Exposed/Infectious/Recovered (SEIR) models pre-coded with both deterministic and stochastic engines.
The parameters in any model are specified in XML configuration files. Users can easily change the weight or significance of various disease vectors (such as the weights of highways, shared borders, airports, etc). Users can also create their own unique vectors for disease. Further details are available in the user manual and design documentation.
The U.S. Map in STEM with Admin 2 resolution showing county adjacency and interstate highways
The original version of STEM was available for downloading on IBM's Alphaworks. It contained easy to follow instructions and many examples (various diseases and maps of the world).
New developers who want to work on STEM II can find useful tools, conventions, and design information in the Welcome Developers article.
The STEM code repository] will be hosted on the Eclipse OHF code repository.
A recent publication on STEM: Ford, D.A., Kaufman, J.H., Eiron, I., "An extensible spatial and temporal epidemiological modeling system", International Journal of Health Geographics 2006, 5:4 http://www.ij-healthgeographics.com/content/5/1/4 (17Jan2006)
About the technology authors
- Daniel Ford, Ph.D., is the former manager of the Web Technologies Department at IBM Almaden and is currently on assignment at the IBM Watson Research Center in New York. He has a wide range of interests and research activities, including software engineering, bioinformatics, and robotics.
- James H. Kaufman, Ph.D., is manager of the Healthcare Informatics project in the Department of Computer Science at the IBM Almaden Research Center. He is also a fellow of the American Physical Society. During his career at IBM Research, Dr. Kaufman has made contributions to several fields, including simulation science and magnetic device technology. His scientific contributions include work on pattern formation, conducting polymers, superconductivity, experimental studies of the Moon Illusion, as well as contributions to distributed computing and grid middleware.
- John Thomas is a Java developer for IBM. He was previously one of the lead programmers for the IBM Almaden TSpaces project and also a member of the OptimalGrid Project at the Almaden Research Center. Mr. Thomas can be reached by e-mail (jthomas119 @ gmail.com)
- Iris Eiron joined IBM Israeli Research Lab in January 1998 and began working at IBM's Almaden Research Center in December 2000. Ms. Eiron's current interests include development and implementation of a national health care information infrastructure.
- Ohad Greenshpan is part of the Healthcare and Life Sciences group in IBM Haifa Research Labs. Mr. Greenshpan is an MSc student for Bioinformatics in Ben-Gurion university, concentrating on Potein Folding algorithms and Structural Bioinformatics. Prior to IBM, Mr. Greenshpan was a member of the Genecards team in Weizmann Institute of Science.