Notice: this Wiki will be going read only early in 2024 and edits will no longer be possible. Please see: https://gitlab.eclipse.org/eclipsefdn/helpdesk/-/wikis/Wiki-shutdown-plan for the plan.
Difference between revisions of "Ebola Models"
(→Model Equations=) |
(→Model Equations) |
||
| Line 38: | Line 38: | ||
The model above also includes clinical isolation in a clinic or hospital (Compartment H) and can be tuned to capture breakdown of infection control (infection between healthcare workers and infected patients). | The model above also includes clinical isolation in a clinic or hospital (Compartment H) and can be tuned to capture breakdown of infection control (infection between healthcare workers and infected patients). | ||
| + | |||
| + | ==='''Basic Reproductive Number'''=== | ||
The basic reproductive number, Ro, is defined as the number of secondary infections caused by one primary infection introduced to a fully susceptible population at a demographic steady state (Anderson and May, 1991; MacDonald, 1957; MacDonald, 1965). From the differential equations above we derive analytically the expression of Ro. | The basic reproductive number, Ro, is defined as the number of secondary infections caused by one primary infection introduced to a fully susceptible population at a demographic steady state (Anderson and May, 1991; MacDonald, 1957; MacDonald, 1965). From the differential equations above we derive analytically the expression of Ro. | ||
[[Image:Ebola_R0.png|500px]] '''The Basic Reproductive Number for Ebola depends on several infection pathways''' | [[Image:Ebola_R0.png|500px]] '''The Basic Reproductive Number for Ebola depends on several infection pathways''' | ||
Revision as of 19:41, 29 September 2014
Other Ebola Models
This page describes a model for Ebola (soon to be) available on this site. This is not the only Ebola model, nor the first on created using STEM. To our knowledge the first STEM Ebola model was created by Dr. Vincent Ruslan from Operon Labs/ His model is slightly different from the one to be release on Eclipse but it's still very informative. Please see it here . You can comment on the model in the forum
Dr. Ruslan is collaborating with Dr Althaus, author of the excellent Sep 2nd, 2014 PLOS paper "Estimating the Reproduction Number of Ebola Virus (EBOV) During the 2014 Outbreak in West Africa". He is affiliated with the Institute of Social and Preventive Medicine (ISPM), University of Bern, Switzerland. Dr Althaus's PLOS Ebola Paper (open access)
Background
The current Ebola outbreak in West African countries may have been triggered by an initial infection in Dec 2013. The first case was thought to be a 2 years old patient lived in a village in Guinea [REF]. This patient, who showed the typical symptoms of the disease (i.e., fever, black stool and vomiting), died on Dec 6 2013. [Ref] A chain of illnesses in patient zero’s family has been found and several family members have caught the infection and died thereafter. While attending the funeral, it is believed that two attendees brought back the virus to their village because funeral tends to bring people in close contact with the body. Contact between mourners and the deceased is a social practice for many African communities and represents and important contact pathway for disease transmission. Clusters of the disease appeared in early 2014 in Guinea, Liberia and Sierra Leone. Ebola Virus Disease (EVD), formerly known as Ebola hemorrhagic fever (EHF) is a fatal illness in humans. Ebola is caused by infection with a virus of the family Filoviridae, genus Ebolavirus. The virus spreads through direct contact with infected blood or body fluids (i.e., urine, saliva, feces, vomit, and semen), contaminated objects (e.g., needles) and infected animals. According to WHO, men who have recovered from the disease can still transmit the virus through their semen for up to 7 weeks after recovery from illness. The incubation period generally ranges between 2 to 21 days (WHO). Symptomatic infection lasts about 10 days. The case fatality rate can be as high as 90%. No specific vaccine or medicine (e.g., antiviral drug) has been proven to be effective against Ebola.
Preliminary Ebola Model
The model described below is a preliminary model the STEM developers are contributing to help researchers get started with their own models. It can easily be extended and as we learn more about the current virus (and as that virus mutates) more sophisticated models will no doubt follow. In particular a future model should separate populations into different demographic groups - those whose practices involve contact with deceased people, and those that do not. The current models uses only one "standard" population so that behaviors is captured only in an average way (that does not vary from country to country). The model below is optimized for the three countries in Africa currently most affected by the epidemic (Guinea, Sierra Leon, Liberia).
Using STEM’s automated model building tool (see Figure 1), we have implemented a preliminary epidemiological model for Ebola based on 8 compartments and 8 differential equations. The code is automatically generated by STEM's model builder and transparent to any user. For more information on the model builder please see the STEM Model Creator page and | this YouTube tutorial.
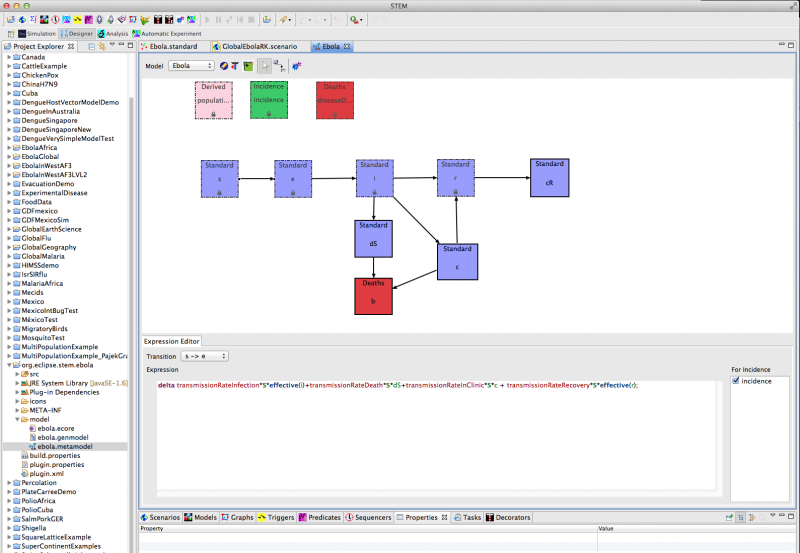 Figure 1: Creating a new model for Ebola using the STEM model builder tool
Figure 1: Creating a new model for Ebola using the STEM model builder tool
The design of the model is shown in Figure 2. It includes the usual Susceptible, Exposed, Infectious, states along with 5 additional compartments: death (but not buried), buried, clinical isolation, and burial (B). The usual R compartment in this model does NOT mean recovered or removed. In this case R represents asymptomatic but still infectious as an STD. WHO indicates transmission, β_r, is still possible for seven weeks after symptoms have gone away through semen by sexual activities among partners. We modeled this post-symptomatic infectious state (R) to capture the possibility of this 7-week of limited transmission. Researchers who question this infection pathway can simple zero out this transmission rate.
Complete recovery is represented by the C compartment. Regarding compartment B, significant infection is believed to happen due to post-mortem contacts with an infectious corpse from the disease, at home or during funeral. This transmission pathway of Ebola is also captured in the model.
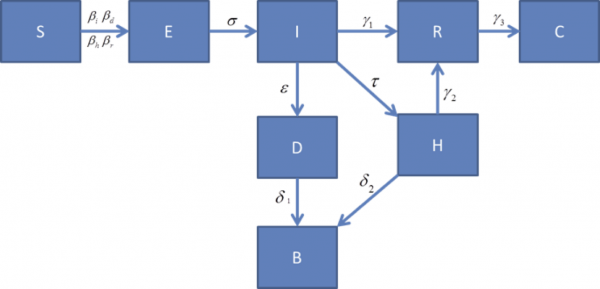 Figure 2: Compartment Model for Ebola
Figure 2: Compartment Model for Ebola
Model Equations
The differential equations defining the model shown in Figure 2 are listed below. If you download the Ebola scenario the terms that build up to form these differential equations are shown in the Model Builder's equation editor - just click on each transition to see the terms. The generated code can be selected in the project explorer (LHS of figure 1).
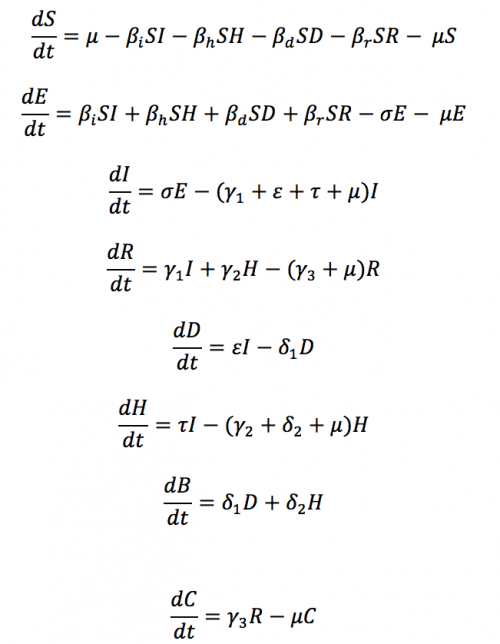 Differential equations defining the compartment model shown in figure 2'
Differential equations defining the compartment model shown in figure 2'
The model above also includes clinical isolation in a clinic or hospital (Compartment H) and can be tuned to capture breakdown of infection control (infection between healthcare workers and infected patients).
Basic Reproductive Number
The basic reproductive number, Ro, is defined as the number of secondary infections caused by one primary infection introduced to a fully susceptible population at a demographic steady state (Anderson and May, 1991; MacDonald, 1957; MacDonald, 1965). From the differential equations above we derive analytically the expression of Ro.
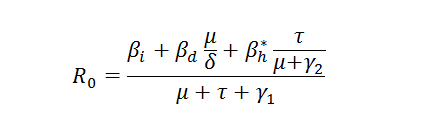 The Basic Reproductive Number for Ebola depends on several infection pathways
The Basic Reproductive Number for Ebola depends on several infection pathways

